My current research focuses on the interpretability and dynamic latent embeddings of deep learning model in temporal data, particularly, multi-view learning and statistical Inference.
My career goal is to develop interpretable large-scale statistical learning methods to advance the science of technology and human medicine.
I believe robust statistical learning methods are necessary and important in handling data heteroscedasticity.
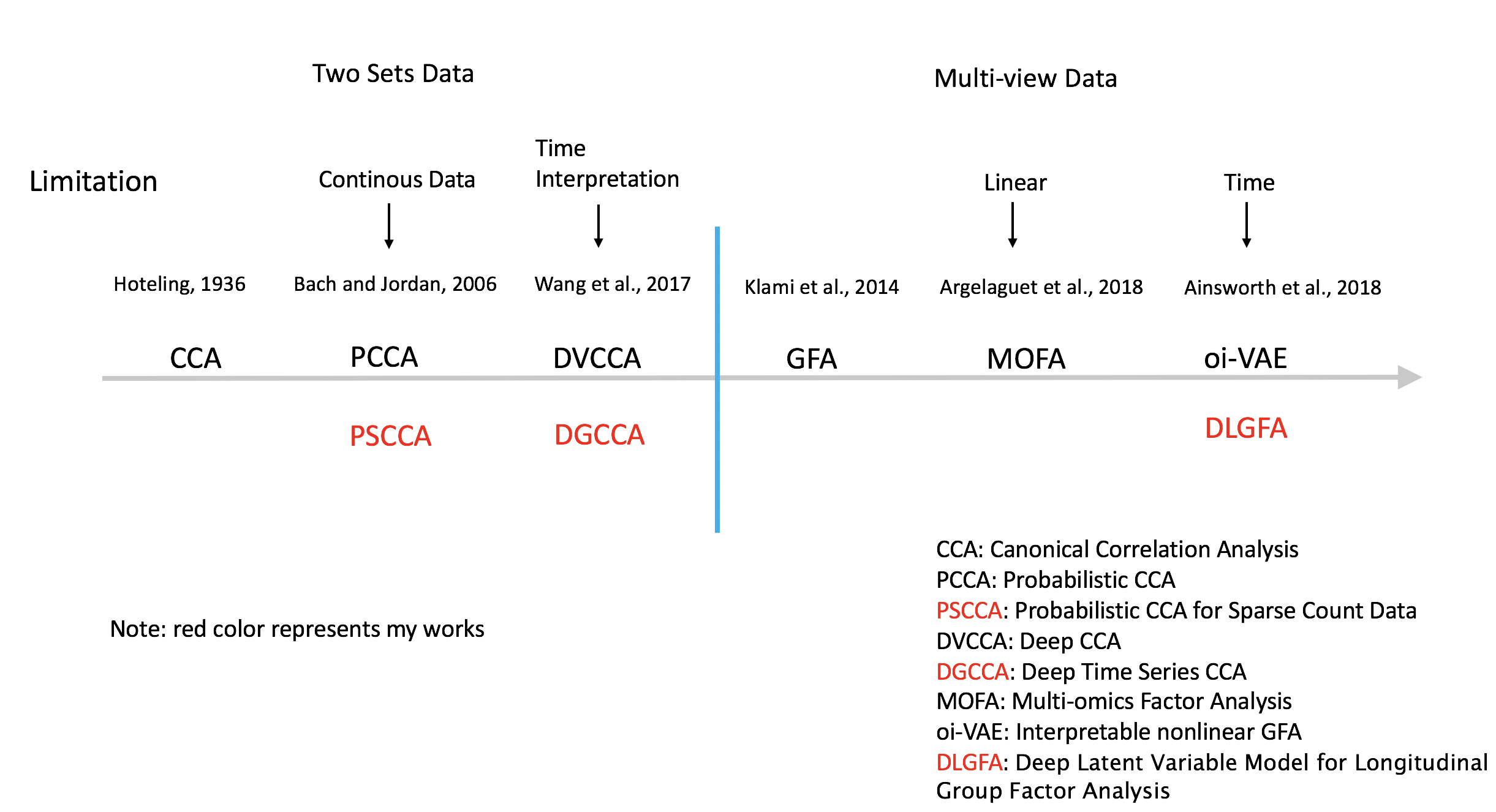
For sequence data in image, voice, or computer vision, by studying the shared and view-specific dynamic latent embeddings of the complex multimodal data will lead to more powerful prediction and interpertable understanding the hidden truths. Like human motion capture, surveillance cameras, and friendship interaction in social media (Facebook, Snapchat, etc.).
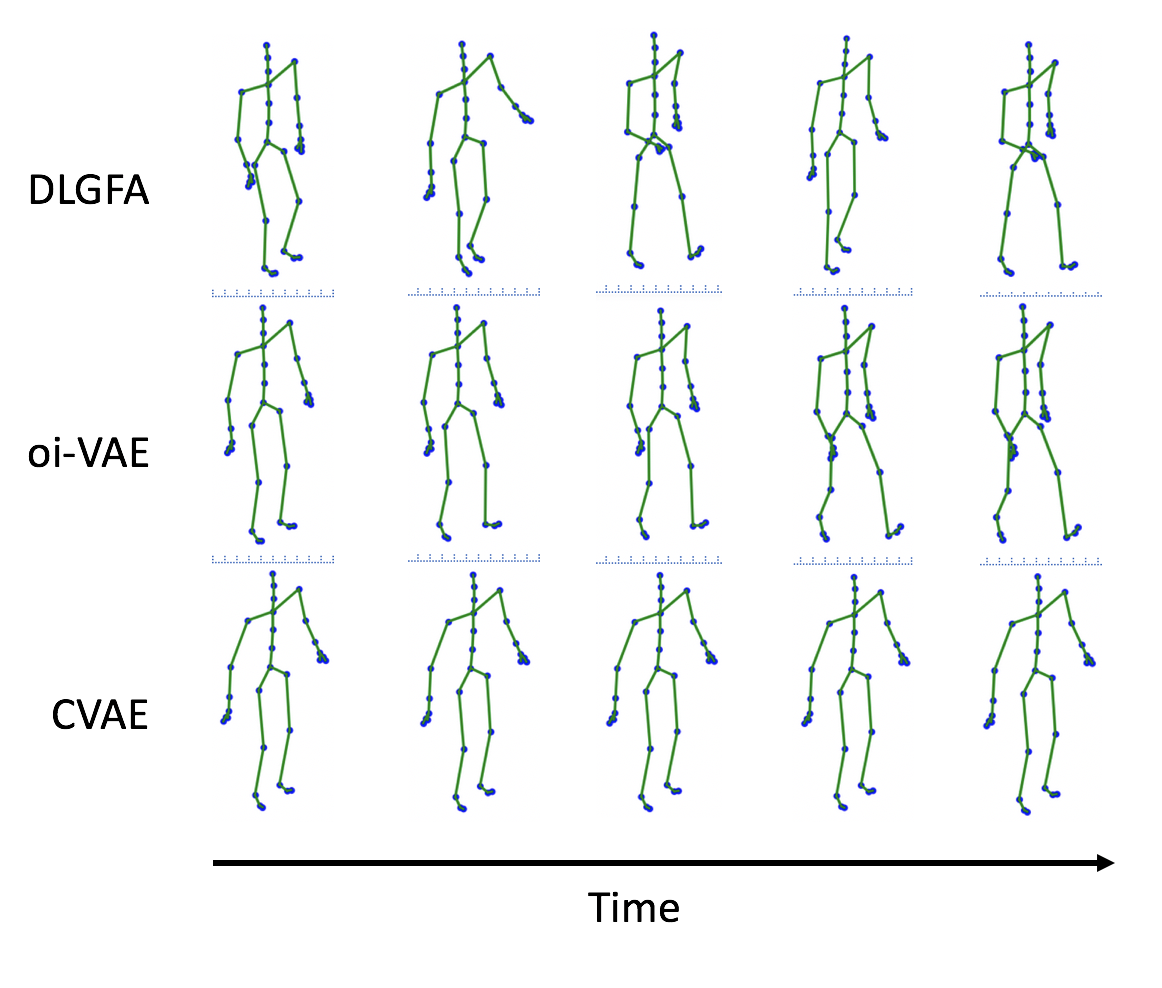
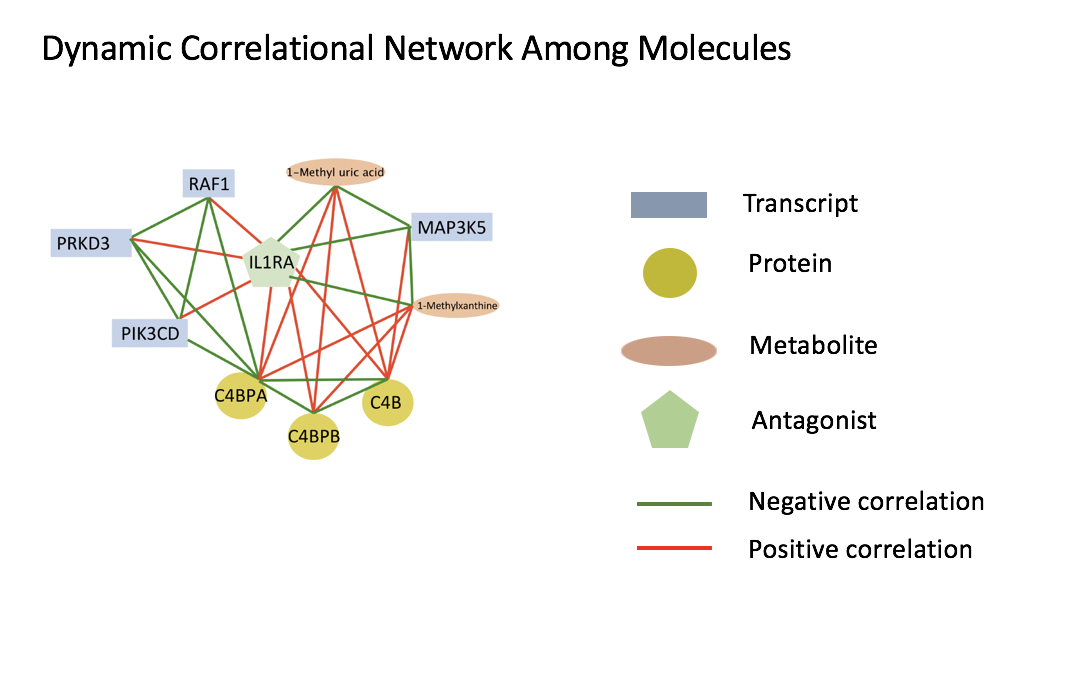
For life science, I develop methods for jointly analyzing multivarite time series data (e.g., transcriptomics, proteins, metabolomics, clinical test, neuroimaging, etc.) and for examining the dynamic dependency among them. To construct the dynamic network with associated gene pathways through deep neural network we can gain meaningful insight for the disease progression.
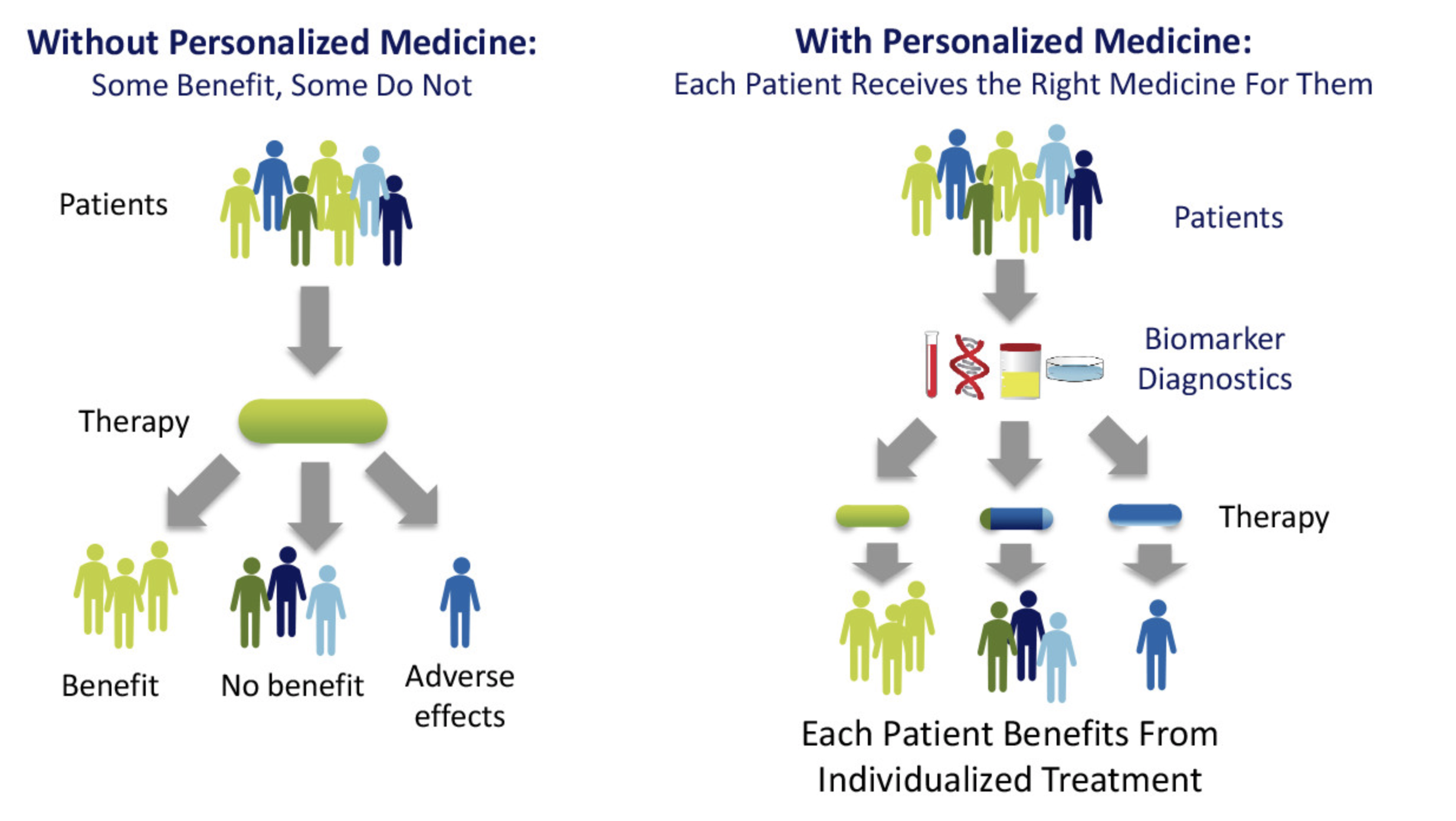
In precision medicine, by examining different subgroups with the maximized treatment effect, we can see a more comprehensive view of the entire population and deliver personalized treatment to cure the rare disease.
Consulting
- Characterize the risk of death after diagnosis from the index cancer vs any other cause, as a function of the disease site and follow up time (Big data around 3.5 million patients). PI: Dr. Zaorsky Nicholas
Existing methods: XGBoost, Deep Neural Network, Random Forest, Logistic Regression, Navie Bayes, etc.
- Survival Analysis on Cancer Patient Clinical Trial Study. PI: Dr. Kathleen Sturgeon
Talks:
-
Deep Interpretable Canonical Correlation Analysis. ISBA 2021, Virtual (07/2021)
-
Deep Latent Variable Model Learning. Snap Research, Santa Monica, CA (09/2020)
-
Interpretable Recurrent Nonlinear Group Factor Analysis. JSM 2020, Statistcal Learning and Data Science Section, Philadelphia, PA (08/2020)
-
Deep Latent Variable Model for Learning Longitudinal Multi-view Data. Genentech, Biostatistics Department, San Francisco, CA (06/2020)
-
Probabilistic Canonical Correlation Analysis for High-dimensional Sparse Count Data[Slides]. ENAR 2020, Count Data: The thought that counts Section, Nashville, TN (03/2020)
-
Summer Internship Project. Penn State Biostatistics Student Seminar, Hershey, PA (12/2019)
-
A Novel Subgroup Identification Based on Exhaustive Search[Slides]. AbbVie SDS Seminar, Worcester, MA (08/2019)
-
Probabilistic Canonical Correlation Analysis for Multiple Groups[Slides]. JSM 2019, Bayesian Statistical Science Section, Denver, Colorado (07/2019)
-
A Bayesian Framework for Rewiring the Topological Network of Intratumoral Cell. ENAR 2018, Atlanta, GA (03/2018)